As the COVID-19 Pandemic began to spread through the U.S. in March, everyone was scrambling. Collectively, we were scrambling to figure out how we could stay safe, manufacture equipment, and get everyone the care they needed. As a geospatial software company, Azavea was scrambling to help the cause by generating an important resource: reliable data.
Coronavirus response policy is necessarily informed by data (this assertion has become a political talking point recently). Elected officials make decisions based on epidemiological models, which are only as good as the data that goes into them. They also make decisions based on capacity, such as tests, or hospital beds and ventilators.
CovidCareMap
Azavea worked with a number of other volunteers to create CovidCareMap: an open source effort to create and visualize a comprehensive national dataset of the hospital system. Our objective was to make the data easy to access and interpret for decision-makers.
Disparate hospital data sources
Before this, one may have assumed that a comprehensive public dataset of all hospitals in the U.S. already existed, and she would be somewhat correct. In fact, Azavea found three datasets with partial information:
- HIFLD (Homeland Infrastructure Foundation-Level Data): Basic information (e.g. location, contact info, number of beds) gathered by the Department of Homeland Security
- HCRIS (Health Care Cost Report Information System): Facility information (e.g. ICU bed/staffed bed capacity) derived from required annual cost reports for all medicare-certified institutions, collected and published by the Centers for Medicare and Medicaid Services
- DH (Definitive Healthcare): A dataset made public by Definitive Healthcare (a healthcare data analytics company), adding information on ventilator and ICU utilization
All three of these datasets combine to offer an exhaustive record of the capacity of every hospital in the country but do not easily relate to each other. There is no standard unique identifier among them, and text fields like the hospital’s name or street address often varied wildly from HIFLD to HCRIS to DH.
One unified dataset
We created a process using both geographic proximity and approximate text matching (comparing name and address fields that may be close but not exactly the same) to join each HIFLD facility to the appropriate records in both the HCRIS and DH datasets. I will discuss our process throughout the rest of the post. You can also follow along using this Python notebook.
We chose to treat the HIFLD facilities as the “authoritative” dataset. This meant that we would try to match each DH and HCRIS facility independently to a hospital in the HIFLD dataset. The DH and HCRIS datasets would never be directly joined to each other, although they would ultimately be paired up via their mutual HIFLD match. This also meant that we would perform a “left” join onto the HIFLD data: each resulting row would have the original attributes from HIFLD as well as the information associated with either DH or HCRIS facility it matched to.
What could go wrong?
Matching facilities from different, unrelated datasets presented many generic matching problems:
- Inconsistent address encoding (e.g. “Road” vs “Rd”)
- Multiple addresses for the same facility (often the case with large medical complexes)
- Outdated data (i.e. one dataset may include an outdated name or address for a hospital that was renamed or moved)
And problems specific to a set of hospital datasets:
- At the margins, the datasets included or excluded slightly different medical facilities (e.g VA health centers, satellite locations, and some outpatient facilities)
- Individuating hospitals from their “systems” or “networks” (i.e. one dataset may consider a network of three health centers to be 1 hospital while another may consider it to be 3)
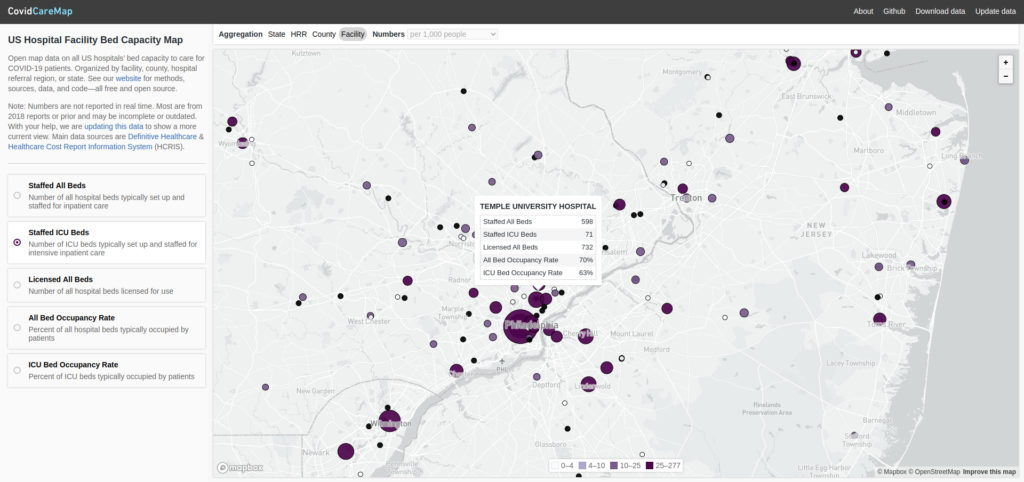
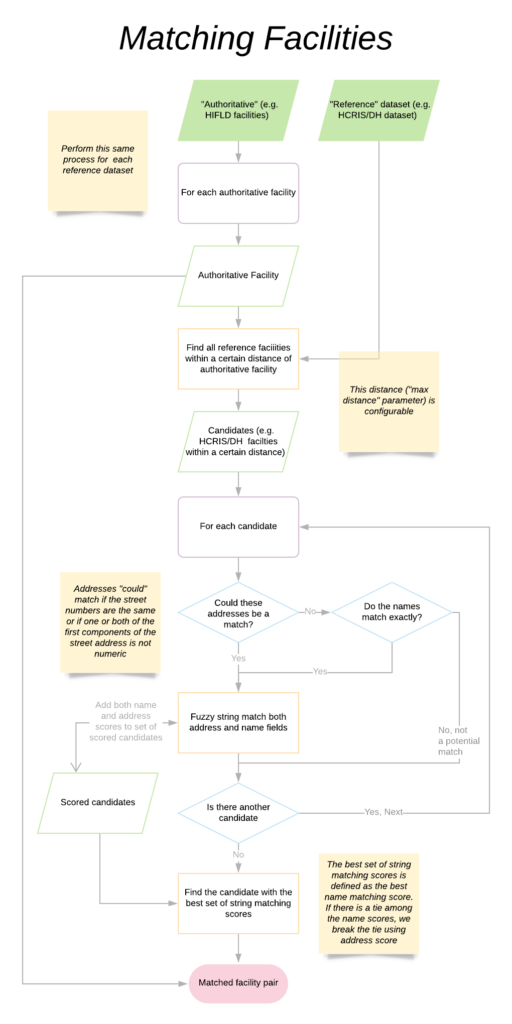
Consider the following example from Buffalo, New York:
The names for the facility are different across all three datasets. The HCRIS name (“KALEIDA HEALTH”) is the name for the healthcare network that includes this hospital. In the second image, you see a children’s hospital that is present in the DH dataset but not in either of the others. The relationship between the hospitals is murky. It’s possible that HIFLD and HCRIS have included the Children’s hospital data in the Buffalo General Medical Center Record while DH does not. We had to build a matching algorithm that would contend with as many of these types of edge cases as possible.
Geographic proximity
The first criterion we used for finding matches between datasets was geographic proximity. If the geographic coordinates for a facility in the HIFLD dataset are 1 mile from facility X and 2 miles from facility Y, it is more likely that facility X is the appropriate match. The logic is straightforward but implementing it requires accurate geographic coordinates for all three datasets.
Geocoding
Of the two non-authoritative datasets (DH and HCRIS), one (DH) came with coordinates attached while the other did not. We used both the Google and Mapbox APIs to geocode HCRIS data. The DH data came with coordinates already attached but upon close examination, we found that many were incorrect or imprecise. Therefore, we passed the DH data through the same geocoding process that we used for HCRIS. In addition to yielding more accurate coordinates for the DH data, this step ensured that we would be using consistently geocoded points for both reference datasets. Our hope was that this would help match consistently from HIFLD to HCRIS and from HIFLD to DH.
Proximity matching
With accurate coordinates for all three datasets, we were able to measure the Euclidean distance between any two candidate points. However, there is more than one way to use proximity when matching points.
Consider trying to match hospital A from the HIFLD dataset to the correct facility in DH:
- Do you simply choose the DH facility that is closest to hospital A? What if the closest point is 100 miles away? This is probably not an accurate match.
- Ok, what about the hospital that is closest, as long as it’s within, let’s say, 25 miles? This could work, but hospital systems are complicated and often clustered together. Let’s say hospital A has three DH facilities within 25 miles: they are 0.2, 0.25, and 18 miles away. Are we so sure that the proximity gap between the first two is great enough to choose #1 over #2?
- So maybe it has to be within 25 miles, the closest match, and be orders of magnitude closer than the next closest? This would probably eliminate some false positives but also increase the number of false negatives (i.e. instances in which we couldn’t find a match but there really was one). After all, the closest option from the previous example may be correct, but it’s just not definitive.
We worked through many different iterations of these rules. Each time we manually checked a portion of the matches to see how accurate it had been.
Ultimately, we settled on using distance as a filtering mechanism. We would specify a maximum distance (500 Meters by default) and use it to reduce the number of candidates from the entire dataset to just those facilities within 500 meters. From there we used string matching to pick from the shortlist of candidates.
Matching text fields
There are many different string fields (e.g. hospital name, hospital type, street address, state, zip code) that describe each hospital in each of the datasets. For the purposes of this task, we selected the two attributes that were most likely to specifically identify a hospital: address (e.g. 123 Main St.), and hospital name.
We created a multi-step process that first screens a hospital’s address for possible equivalency then compares the names contingent on the results of the address screening. We considered hospital addresses “possible matches” if they had the same street number. Consider the following two addresses:
- 2700 Riverview Drive
- 2700 N Riverview Dr.
It is entirely possible that these two address strings refer to the same hospital location. We found that there was more variance in street name encoding than in building numbers. Therefore, we used building numbers as our first-pass filter criteria. These two addresses would be considered a “possible match.”
We also considered pairs to be potential matches if we could not parse the first components of one or both of the addresses to integers. This would be the case with either of the following versions of the example address:
- 2700-2900 Riverview Dr.
- 27th St. and Riverview Dr.
These addresses could both be correct interpretations of the first set. Therefore, we decided to consider pairs that included similar addresses in case they did turn out to be correct matches.
Fuzzy string matching
After the screening process, we used fuzzy string matching to compare both address and hospital names. This method allowed us to compare fields that were often similar but not exactly the same. For this task, we used a python library called rapidfuzz, which assigns numeric similarity scores to pairs of strings.
Finally, we determined the best match among the “possible” candidates using a combination of the name and address scores. We chose the candidate with the most similar name to the reference facility. If the top-scoring name matches had the same score, we used the address score as a tie-breaker.
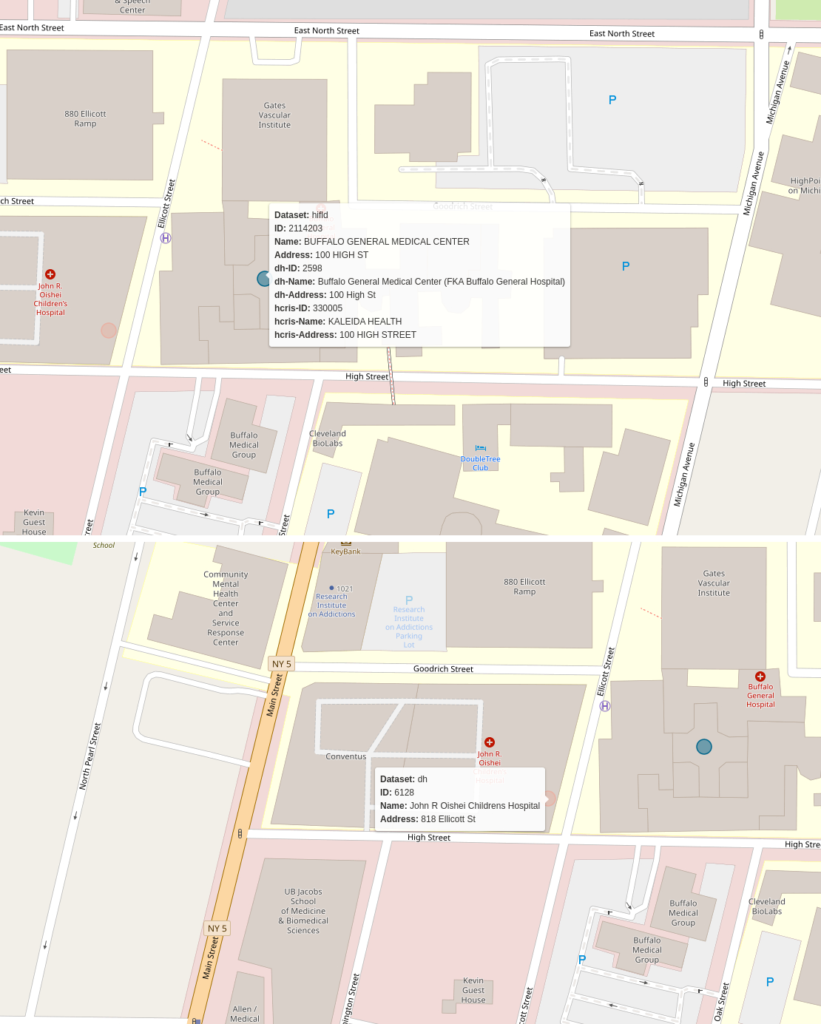
Putting it all together
We can summarize the whole process with the following diagram:
In practice, we matched both the DH and HCRIS facilities independently to HIFLD at the same time. Of course we had to keep track of which HCRIS and DH facilities we had already matched to HIFLD. This presented an additional engineering challenge but ensured that we were not double-counting facilities that could be considered the best match for multiple hospitals within the authoritative set.
In the end, we were able to make great strides in combining the datasets. We can not provide an exact accuracy measure without manually checking the 6,600 facilities of the final dataset. However, we correctly identified many “difficult” matches (e.g. pairs of facilities with one outdated location, address, or name).
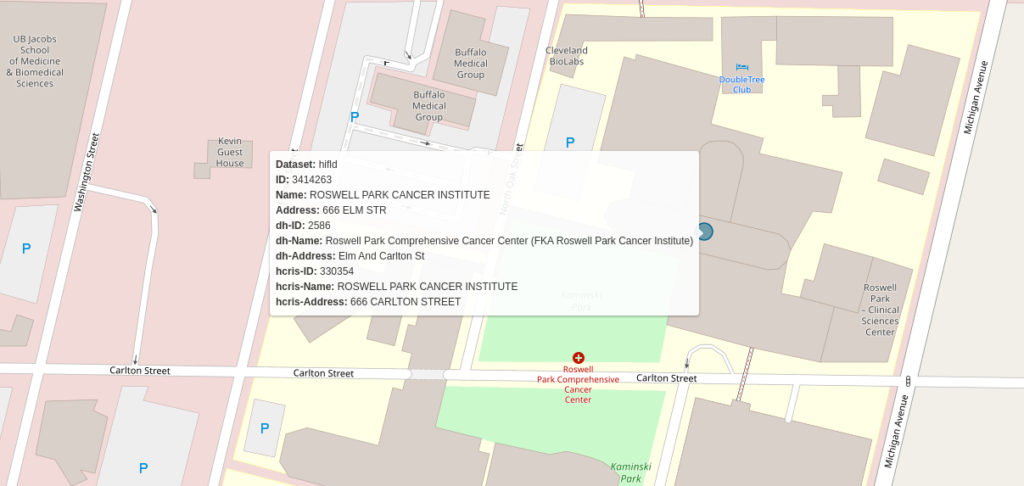
Check out a visualization of these data at CovidCareMap.org. Download these data on the project’s github page.
We hope that by sharing this methodology, Azavea can help decision-makers and researchers who require a comprehensive national dataset of the hospital system.
